Other Research Projects
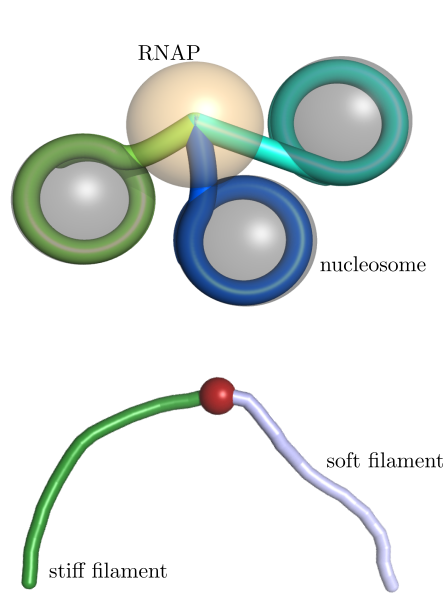
Kinetics of mechanosensitive molecular processes
Effect of biofilament fluctuations on enzymatic rates
Many enzymatically driven biomolecular processes have rates that are mechanosensitive -- modulated by applied force. Inside a living cell, the forces experienced by an enzyme or molecule can exhibit large fluctuations over time. As a consequence, the reaction rate may deviate substantially from that expected at a given average force, and molecular interactions or constraints which control fluctuations can have important effects on reaction kinetics.
We have explored two biologically important systems where force fluctuations can play an important role. The first of these is transcription by RNA polymerase when it encounters the nucleosomes that form a basal level of packaging for the eukaryotic genome. As the polymerase unpeels the DNA from a nucleosome, it encounters increasing yet fluctuating forces that can trigger pausing and modulate its overall transcription rate. Our quantiative model of this phenomenon has highlighted the key role played by unpeeling fluctuations of nucleosomal DNA. The model has been leveraged to link single-molecule experimental data on nucleosome-DNA binding affinity and observations of polymerase pausing at specific basepair regions as it enters the nucleosome. This work was carried out in collaboration with the Bustamante group at UC Berkeley.
Fluctuations of cytoskeletal filaments can also play an important role in modulating their interactions with the enzymes that drive filament dynamics. Actin filament turnover relies on the cofactor cofilin, which coats regions of a filament and triggers severing at the boundaries between coated and uncoated regions. Cofilin is known to both undertwist and soften the actin filament upon binding. Using analytical models grounded in the statistical mechanics of semiflexible filaments, we have shown that copolymers with soft and stiff regions can experience enhanced thermal fracture rates at the junction points. This general result points to a purely mechanical mechanism by which cofilin may contribute to actin fracture and severing.
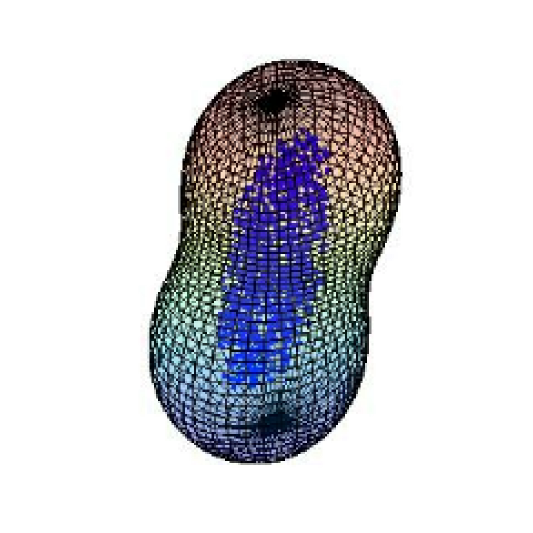
Particle Mixing in a Deforming Domain
Cytoplasmic mixing through cell shape deformation in motile neutrophil-like cells
The motion of intracellular particles can be driven not only by diffusion and active motor-based transport but also by the bulk cytoplasmic flows arising from gross deformation of the cell. A general problem of interest is to quantify the effect of active fluctuations of the cellular membrane on transport and encounter rates of particles within the cell. Using a spectral approach to model fluctuating quasi-spherical vesicles, we demonstrate that shape deformations of the vesicle can lead to overall particle motion that dominates over diffusion on second to minute timescales, speeding up the rate at which particles encounter different regions of the fluid domain.
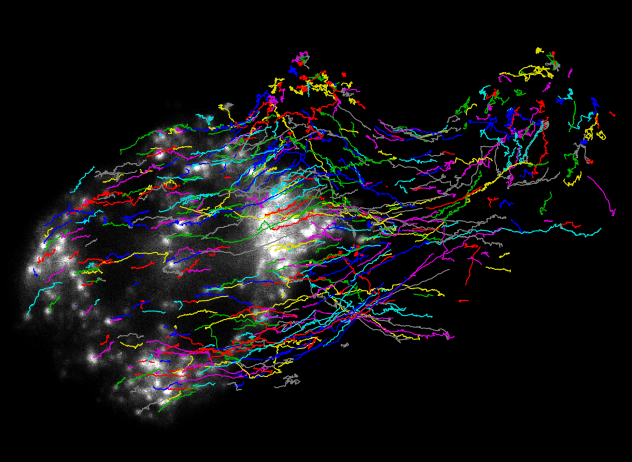
Analyzing Rheology in the Presence of Flow
Statistical methods for extracting mechanical properties from particle trajectories in a flowing medium.
Tracking the motion of single particles provides valuable information about the material environment in which they are embedded. However, in the case of highly dynamic cells, the stochastic motion of the particles is confounded by large-scale cytoplasmic fluid flow. We have developed a novel technique for analyzing particle tracking data in the presence of flow. The method uses wavelets to separate trajectories into a smoothened slowly-varying component and a "noise" component encompassing Brownian (or fractional Brownian) motion. It has been used to demonstrate that the cytoplasm of crawling neutrophils behaves primarily as a viscous fluid, with rheological properties that can be grossly altered by drug treatments of the cells.

Cell Wall Mechanics in Bacterial Cell Division
Stress and fracture in the separation of Staphylococcus aureus daughter cells
Staphylococcus aureus bacteria grow and divide in a discrete fashion, with extremely fast popping events splitting one cell into two. I investigate the physics underlying such popping events by modelling the bacterial cell wall as an elastic shell, with stresses arising from a combination of turgor pressure and differential growth of the shell. Additionally, treating the peripheral band surrounding the shell as an expanding ductile ring helps explain the formation of periodic defects in the cell wall prior to division.
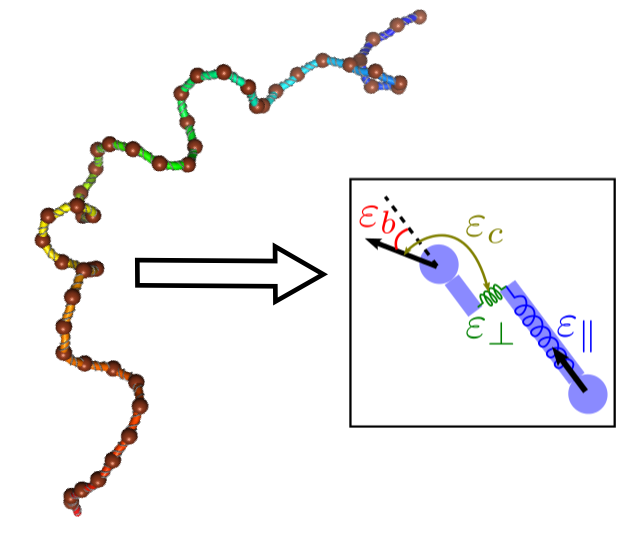
Systematic Coarse-graining of Polymer Models
Statistical mechanics and renormalization of general polymer models to effective semiflexible chains at long lengths
At long lengths, polymer chains exhibit general physical properties that are independent of their detailed structure. I developed a systematic approach for mapping polymer chains to a coarse-grained model called the "shearable worm-like chain" which can reproduce the statistics of the chain at intermediate to long lengths. This chain model is analytically tractable and suitable for simulations, where the coarse-graining length scale can be tuned to suit the application of interest.

Chromatin Fiber Structure
Mapping the energy landscape of regular helical structures for the 30nm chromatin fiber
The eukaryotic genome is packaged into a hierarchical structure that involves wrapping the DNA into periodic nucleosome structures, that are then packed into regular helical chromatin fibers. We consider the role of DNA mechanics in these helical fibers. Treating the linker DNA as a wormlike chain and focusing on regular helical fibers, we perform geometry optimization to characterize the possible low-energy fiber structures. Our results indicate a multitude of possible helices, indicating that there is unlikely to be a single thermodynamically determined structure for the chromatin fiber.
Target Search via Multimodal Passive Transport
Dynamics of DNA binding proteins in searching for target sites on the genome
DNA binding proteins find their target sites by a combination of 3D diffusion and 1D sliding along nonspecific DNA sequence. Using analytic and simulated reaction-diffusion models, we explored the kinetics of this process, focusing on the role of DNA configuration and confinement. We find that for the parameter regime relevant to in vivo systems, the complex facilitated diffusion process can be represented as an effective diffusion to an antenna region around the target site.