The Koslover Group Department of Physics, UCSD
Transport and Kinetics in the Intracellular World
The Koslover group is focused on physical modeling of eukaryotic cells. Many of the questions we address center on intracellular dynamics and transport -- how do different components, ranging from molecules to organelles, navigate the complex world inside a living cell? How do interactions between organelles contribute to distributing and dispersing cellular components? How do large-scale structures (eg: reticulated organelle networks) emerge from smaller-scale dynamic processes? To address these questions, we use a combination of pencil-and-paper theoretical modeling together with meso-scale computational simulations that aim to establish the key physical principles and parameters that govern intracellular dynamics. We work closely with experimental collaborators to compare model predictions against data from live cells.
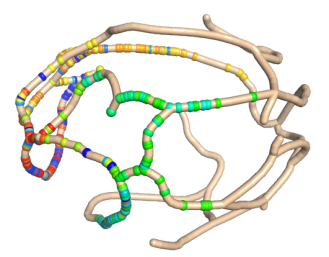
Simulated trajectory of a diffusion particle on a yeast mitochondrial network.
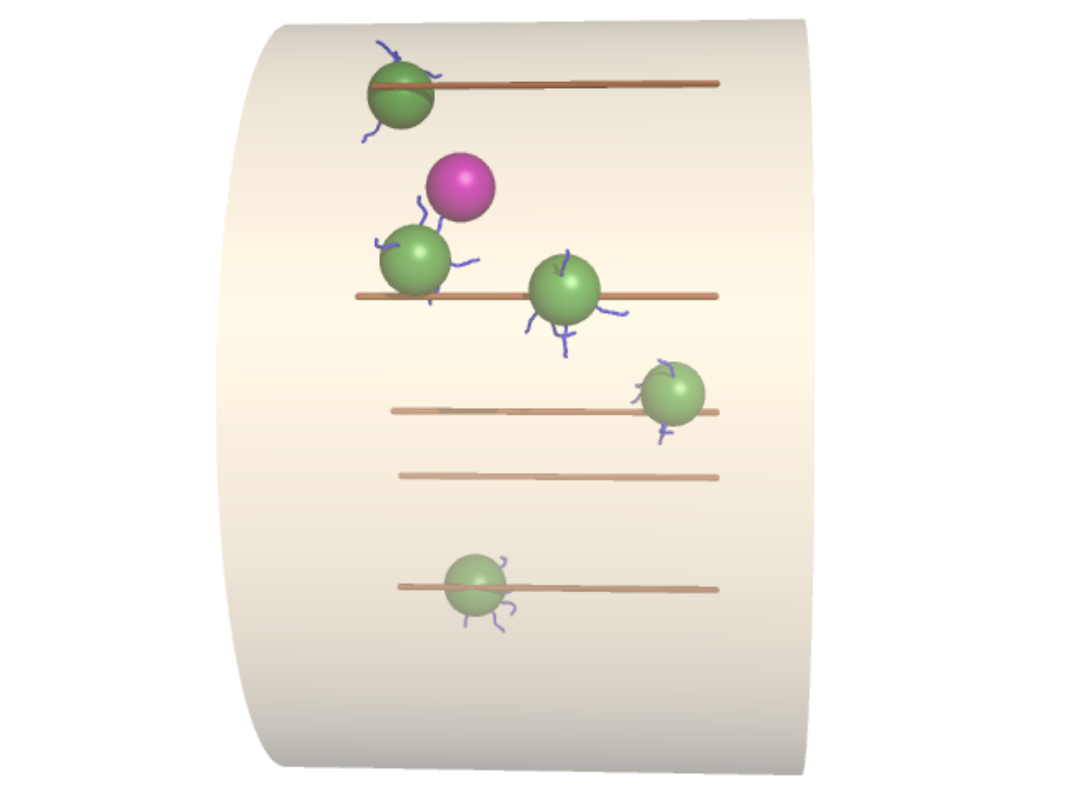
Model for initiation of peroxisome transport via hitchhiking on moving endosomes in a fungal hypha.
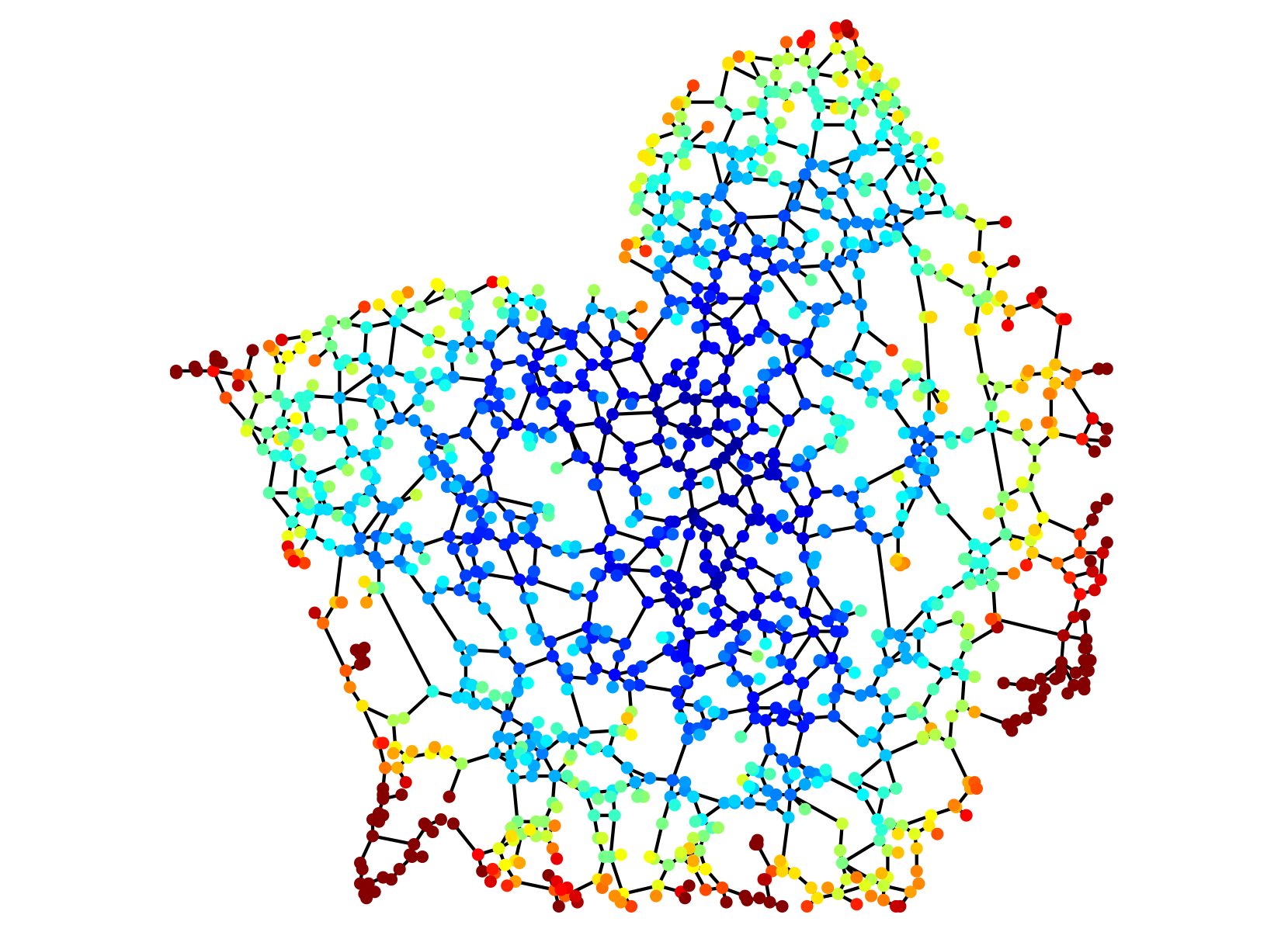
Diffusive search times on network structure for a portion of the peripheral endoplasmic reticulum in a mammalian cell.